Bringing the power of generative AI across the cancer continuum
Experience the future of cancer diagnosis with Paige, today.
Our innovative AI technologies, built on the largest datasets globally, are already transforming pathology and making generative AI accessible to doctors.
The Paige Platform allows for full digitization of the pathology laboratory to elevate doctor’s capabilities beyond what is possible with traditional pathology and drive workflow efficiency and AI driven precision in cancer.
Paige’s generative AI is now positioned to unlock the deepest secrets of cancer and usher in the next generation of diagnostic tools that we believe will save lives.
Trusted Paige Customers Include
We empower doctors today with the tools to shape a better tomorrow
clinician utilization of paige
With an installation base spanning across 5 continents, Paige is trusted and utilized by renowned hospitals and laboratories around the world as part of their clinical workflows









A New Standard For Hospitals & Labs
Paige products have defined a new standard in the field of pathology. Our powerful digital platform and robust portfolio of cutting-edge AI applications streamline pathology workflows and provide actionable diagnostic insights to directly inform patient care.
The scalable, AI-ready foundation for digital pathology
Paige’s portfolio of clinic-ready AI applications support pathologists in their detection & diagnosis of cancer
AI applications for the detection of molecular biomarkers on digitized tissue images not typically visible to the eye
Independently validated and clinically utilized by labs around the world
negative predictive value3
70%
reduction in cancer detection errors4
65.5%
reduction in time to diagnosis3
39%
reduction in second opinion requests1
21%
reduction in IHC requests1
Experience The future of pathology
Transforming the way pathologists work through a comprehensive digital pathology platform and robust AI applications. With interoperability and AI integration at the core of all Paige products, Paige represents the only solution labs need for precise, confident diagnosis today and in the future.
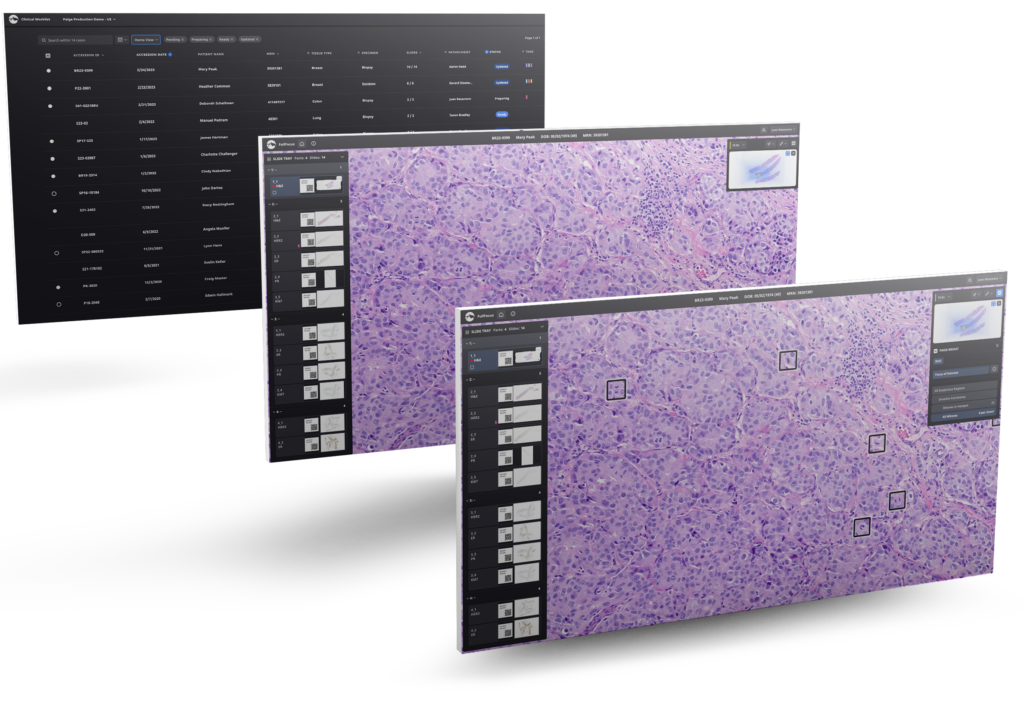
The Standard for AI in Pathology
As the first and only company to receive FDA approval for an AI application in pathology, Paige has set the standard of excellence for developing cutting-edge applications that excel in identifying cancer features solely from H&E-stained whole slide images.
Cutting-Edge Technology
Paige is the global leader in generative AI for pathology, leveraging exclusive access to the largest and most diverse dataset in cancer diagnostics to build cutting-edge applications.
Unrivaled Expertise
Our extensive experience assisting both large and small labs with digitization allows us to guide customers through the complexities of digital pathology, ensuring a seamless transition and fulfillment of digital pathology and AI’s full potential. Supported by dedicated customer success teams worldwide, Paige remains committed to day-to-day support to ensure labs are set up for success today and in the future.
Our AI solutions provide a clear path into the future of pathology.
industry partnerships
Through strategic collaborations with industry leaders, Paige has established professional partnerships that propel the advancement of digital pathology.









Work with Paige
Talk to us
Get an overview of Paige and our current offering by scheduling a short demonstration with a member of our team
Put us to the test
Upload your own digital images to see how FullFocus® and our AI applications could work at your lab
Ready to connect?
Let's talk about how Paige can help you bring confidence and accuracy to every diagnosis.
- 1Eloy C, Marques A, Pinto J, et al. Artificial intelligence–assisted cancer diagnosis improves the efficiency of pathologists in prostatic biopsies. Virchows Arch. 2023. doi: 10.1007/s00428-023-03518-5
- 3da Silva LM, Pereira EM, Salles PGO, et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J Pathol. 2021;254(2):147–158. doi: 10.1002/path.5662
- 4Raciti P, Sue J, Retamero JA, et al. Clinical Validation of Artificial Intelligence–Augmented Pathology Diagnosis Demonstrates Significant Gains in Diagnostic Accuracy in Prostate Cancer Detection [published online ahead of print, 2022 Dec 20]. Arch Pathol Lab Med. 2022. doi: 10.




